
Cluster analysis
- Cluster analysis: numerical procedure used to form groups
of
entities in some specified manner
- Cluster analysis represents an attempt to find structure
- assumes that group structure exists, but that classes
(groups) are unspecified
- Ability to detect group structure in data depends on:
- underlying structure, and
- technique used
- Terminology:
- Hierarchical: optimizes route along which structure is
sought
- Nonhierarchical: optimizes some property of the
group
being formed
- e.g., consider the following
data:
- Minimizing the increase in SS between groups is a route-based
(i.e., hierarchical) technique. Adding quadrat * to group 1 -->
incr. in SS of 50, and adding quadrat # to group 2 --> incr. in
SS of 100; add * to group 1 based on optimization of route. If
the decision is based on SS of groups themselves (vs. incr. in
SS), we would be measuring a property of the group (i.e.,
nonhierarchical)
- Divisive: group is repeatedly halved
- Agglomerative: each sampling unit starts by itself, then
group is formed from most similar pair of
entities [and so on]
- Monothetic: decision rule is based on one particular
characteristic (e.g., presence of sp. A -->
group 1; sp. A absent --> group 2)
- Polythetic: decision rule takes all variables (species)
into account
- We will focus on polythetic agglomerative hierarchical methods
- Considerations for all methods:
- What measure is used to indicate similarity (or
dissimilarity) between entities?
- How is this measure used?
- Consider the following data:
| Quadrat | Species A | Species B |
| 1 | 15 | 9 |
| 2 | 12 | 8 |
| 3 | 17 | 13 |
| 4 | 0 | 7 |
| 5 | 8 | 0 |
| 6 | 3 | 12 |
- We can plot quadrats in species-dimensional
space
- Single-linkage clustering (syn. nearest-neighbor
clustering)
- Responses to considerations:
- Euclidean distance (i.e., distance between points)
is
the measure of similarity
- Distance between 2 entities (quadrats, clusters) is
defined to be the shortest distance involved in the
comparison
- Euclidean distance between quadrats:
- d(j,k) =
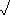 [
[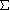 (Nij
- Nik)2]
(Nij
- Nik)2]
| where d(j,k) = | distance between j and k, |
| p = | number of species, |
| i = | species, |
| j = | quadrat, and |
| k = | quadrat |
- e.g., d(1,2) =
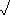 [(15-12)2
+ (9-8)2] =
[(15-12)2
+ (9-8)2] = 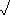 10 = 3.16
10 = 3.16
- All possible distances:
| | 1 | 2 | 3 | 4 | 5 | 6 |
| 1 | - | 3.16 | 4.47 | 15.16 | 11.40 | 12.32 |
| 2 | | - | 7.07 | 12.04 | 8.94 | 9.85 |
| 3 | | | - | 18.03 | 15.81 | 14.04 |
| 4 | | | | - | 10.63 | 5.83 |
| 5 | | | | | - | 13.00 |
- Smallest possible number in this matrix is 3.16 --> quadrats
1 and 2 are most similar (i.e., they have shortest euclidean
distance) among all paris; therefore, quadrats 1 and 2 form a
group
- All possible distances of entities (quadrats + 2-quadrat
cluster):
| | 1,2 | 3 | 4 | 5 | 6 |
| 1,2 | - | 4.47 | 12.04 | 8.94 | 9.85 |
| 3 | | - | 18.03 | 15.81 | 14.04 |
| 4 | | | - | 10.63 | 5.83 |
| 5 | | | | - | 13.00 |
- Smallest number in this matrix is 4.47 --> quadrat 3 joins
the previously-formed group
- All possible distances of entities:
| | 1,2,3 | 4 | 5 | 6 |
| 1,2,3 | - | 12.04 | 8.94 | 9.85 |
| 4 | | - | 10.63 | 5.83 |
| 5 | | | - | 13.00 |
- Smallest number in this matrix is 5.83 --> quadrats 4 and 6
join to form a new group
- All possible distances of entities:
| | 1,2,3 | 4,6 | 5 |
| 1,2,3 | - | 9.85 | 8.94 |
| 4,6 | | - | 10.63 |
- Smallest number in this matrix is 8.94 --> quadrat 5 joins
group comprised of quadrats 1,2,3
- All possible distances of entities:
- Final step is joining together (clustering) of both groups
- Dendograms are usu. constructed to provide a simple visual
summary of cluster analysis steps
- Single-linkage clustering:
- oldest method (obsolete for ecological data)
- "space-contracting"--as a group grows, it becomes more
similar to other groups, leading to "chaining"
- properties of group are ignored--individual quadrats
are compared
- Complete linkage (furthest neighbor) clustering
- Identical to single linkage clustering except that the
distance between entities is defined as the point of maximum
distance
- e.g., distances:
| | 1 | 2 | 3 | 4 | 5 | 6 |
| 1 | - | 3.16 | 4.47 | 15.16 | 11.40 | 12.32 |
| 2 | | - | 7.07 | 12.04 | 8.94 | 9.85 |
- All distances of entities after quadrats 1 and 2 are joined:
| | 1,2 | 3 | 4 | 5 | 6 |
| 1,2 | - | 7.07 | 15.16 | 11.40 | 12.32 |
| i.e., | d(1,3)
= 4.47 |
| | d(2,3)
= 7.07; |
| thus, | d[(1,2),3] = 7.07 |
- w/ single linkage clustering, dist = minimum distance -
-> d[(1,2),3] = 4.47
- Decision rule is still based on smallest distance, but
distances are calculated differently
- Characteristics of complete linkage clustering:
- "space-dilating"--as a cluster grows it tends to become
more dissimilar to others --> non-chaining
- group structure is ignored; as w/ single linkage
clustering, comparisons are based on indiv. quadrats
- results often similar to "minimum-variance" clustering
- Centroid clustering
- Distance between 2 clusters is defined as the euclidean
distance between their centroids
- Two groups are joined if the distance between their
centroids is the smallest of all possible "choices"
- e.g., distance between groups:
- To calculate centroid:
| Quadrat | Species A | Species
B |
| 1 | 15 | 9 |
| 2 | 12 | 8 |
| 3 | 17 | 13 |
| 4 | 0 | 7 |
| 5 | 8 | 0 |
| 6 | 3 | 12 |
- First step is identical to single linkage clustering, since
groups are single quadrats. After quadrats 1 and 2 are
joined, centroid(1,2) = [(15+12)/2, (9+8)/2] = (13.5,8.5).
- centroid (1,2,3) = [(15+12+17)/3, (9+8+13)/3] =
(14.333,10)
- Then euclidean distance is calculated not between
nearest quadrats in group (single linkage) and not
between furthest quadrats in group (complete linkage),
but between centroids of groups
- Thus, group structure is used in determining between-
group similarities
- A disadvantage of centroid clustering is the potential for
reversals
- After a fusion, the next fusion occurs at a less
dissimilar point (i.e., closer distance)
- e.g., consider these 3 quadrats w/ 2 species:
| Quadrat | Species A | Species B |
| 1 | 26 | 10 |
| 2 | 34 | 6 |
| 3 | 34 | 15 |
- d(1,2) =
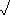 [(26-34)2 + (10-6)2] = 8.944
[(26-34)2 + (10-6)2] = 8.944
- d(1,3) =
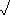 [(26-34)2 + (10-15)2] = 9.434
[(26-34)2 + (10-15)2] = 9.434
- d(2,3) =
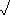 [(34-34)2 + (6-15)2] = 9.000
[(34-34)2 + (6-15)2] = 9.000
- Quadrats 1 and 2 are joined --> centroid =
[(26+34)/2,(10+6)/2] = (30,8)
- d[(1,2),3] =
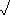 [(30-34)2 + (8-15)2] = 8.062
[(30-34)2 + (8-15)2] = 8.062
- Thus, the second fusion occurs at a smaller
distance than the first fusion (i.e., this
indicates these entities are more similar than
those joined by the first
fusion):
- Centroid clustering incorporates information about the group
when joining groups (vs. single linkage and complete linkage
clustering, which do not)
- However, reversals create interpretational difficulties, and
this has discouraged widespread use of clustering techniques
which have potential to show reversals
- Comparison studies have shown that single linkage and
centroid clustering behave similarly
- Minimum-variance clustering (Ward's method) (syn. Orloci's method
in ecological literature)
- Concept: we can measure the sum of the distances2 of
the
members of a group from the group centroid as an
indicator of group heterogeneity or dispersion
- Distance (similarity) measure: euclidean distance
- Fusion rule: groups are joined only if the increase in
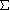 d2
is less for that pair of groups than for any
other pair
d2
is less for that pair of groups than for any
other pair
- Ward's method lends itself to a measure of "classification
efficiency":
- SStotal =
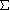 d2
of all quadrats from centroid
d2
of all quadrats from centroid
- At any point in the analysis, SS can be calculated for
each group (i.e., within-group heterogeneity or
dispersion)
- Thus, a percentage can be calculated which
indicates the proportion of total variability
explained by each group:
SSgroup/SStotal
- Characteristics of Ward's method
- Minimizes dispersion within groups
- Like complete linkage clustering, it favors the
formation of small clusters of approximately equal size
- Incorporates information about groups, not merely about
individual quadrats
- Computationally complex and time-consuming compared to
other methods we've discussed
- Widely applied in ecology, especially recently (since
computers have overcome problems w/ computational
complexity)
TWINSPAN
Graphics
To this point, we have discussed only euclidean distance as a
measure of similarity
- Another measure of similarity is chord distances
- Chord distance is still a linear measure of distance, but a
spatial transformation is imposed
- Points falling short of chord are stretched, those beyond
chord are shrunk
- Distance between entities is measured along the arc
(geodesic distance) or across arc, on straight line
- Disadvantages:
- use of chord distance implicitly assumes species are
independent
- in practice, using Ward's method w/ chord distance
causes "chaining"
- For these reasons, chord distances are not widely used
- TWINSPAN (Two Way INdicator SPecies ANalysis)
- Unlike the other clustering methods we have discussed, TWINSPAN
is a divisive method
- TWINSPAN algorithm
- Samples are ordinated w/ RA
- A crude dichotomy is formed: the RA centroid is used
as a dividing line between two groups (negative and
positive)
- The dichotomy is refined by a process comparable to
iterative character weighting
- Dichotomies are ordered so that similar clusters are
near each other
- Assuming higher groups have already been ordered,
ordering of lower groups proceeds by taking into
accound similarity of nearby clusters
- In the absence of ordering, arrangement of
groups 1-8 is arbitrary (i.e., hierarchical
structure only indicates that 1 should be
next to 2, 3 next to 4, etc.)
- Ordering places 2 next to 3 if 2 is more
similar to B than 1 and 4 is more similar to
C than 3
- For this reason, it is said that the
dichotomy is determined by relatively large
groups, so that it depends on general
relations more than on accidental
observations
- Species are classified
In light of quadrat classification: based on fidelity
to particular sites (clusters of quadrats)
- Structured table is made from both classifications
- Presentation of cluster analysis results
- Species-by-site (i.e., species-by-quadrat) table used by
TWINSPAN
- Additional information is usually given for sites
(quadrats)--e.g., environmental variables
- Rarely used w/ large data sets because table is very
large and patterns are not readily discernible
- Dendogram
- Combined w/ ordination (clusters or communities shown on
ordination diagram)
- Interpretation of cluster analysis results
- Calculate descriptive statistics at each dichotomy
- Perform discriminant analysis at each dichotomy to quickly
identify environmental variables which are "most different"
between clusters
- Statistical tests of significance may be used to determine
whether environmental variables are "different" on each side
of a dichotomy
Discriminant analysis
Used to assign members to groups, or differentiate between groups
Groups are pre-determined
Assumes existence of a well-defined group structure; also assumes
data are multivariate normally distributed
Consider a plot of quadrats in species-dimensional
space
- Conceptually, discriminant analysis
seeks similar entities and
groups them together.
- Similarity is sought by using Mahalanobis distance (a
distance measure which "corrects" for the correlation
between species). It is identical to euclidean distance if
species are uncorrelated.
- Use of Mahalanobis distances requires multivariate
normality
- Unlike univariate ANOVA, discriminant analysis is
sensitive to deviations from multivariate normality
- DA is a maximization technique: it does an F-test to
determine which variable maximally discriminates between
groups
- Mechanically, the procedure uses prediction equations
- equations are called classification or discriminant
functions
- they are analogous to regression equations
- e.g., R = c1(X1) +
c2(X2) + c3(X3) + ... +
cn(Xn), where
- R = biogeographic province
- c = constant
- X = species abundance, for various species (Xi's)
- Values of R indicate province to which quadrats should
be assigned
- Coefficients, then, determine to which group a quadrat will
be assigned
- Coefficients are sensitive to deviations from
multivariate normality
- Coefficients are sensitive to prior probabilities
(which must be assigned)
- DA assumes that covariances are equal within groups
- But if group structure exists, then relationships between
species should be different in different groups
- Finally, there is a problem w/ statistical bias:
- Group membership is based on all samples, including the
sample being assigned to a group. Thus, the quadrat being
considered for group membership contributes info to the
determination of coefficients.
- DA is often used in ecology, at least partially because it is one
of few multivariate tools which allows a statistical test of
hypothesis
- In practice w/ real data, calculation of the F-statistic is
so sensitive to violations of assumptions (bias, normality,
equality of covariances, assignment of prior probabilities)
that the F-test is badly flawed
- Nonetheless, discrimination is often quite good--as a
maximization technique, DA capitalizes on small
discriminating differences --> good classification into
groups
- Furthermore, R2 values are usually high, because sample
size (number of quadrats) is usually small relative to
the number of independent variables (species)
- Therefore, DA almost always looks good on paper
- However, the associated F-test should be viewed w/
extreme caution
- Williams (1983, Ecology 64:1283-1291) concluded that there is
widespread use of DA in ecology, and almost always it is done
incorrectly
- He did not recommend using DA only when assumptions are
rigorously satisfied
- But there is a difference between "exploration" and
"confirmation"
- Statistical procedures can be used to explore data
whether assumptions are met or not
- But any perceived patterns should be regarded as
preliminary and should be used to suggest hypotheses
which can be subsequently tested
Previous
lectureNext
lecture
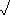 [
[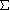 (Nij
- Nik)2]
(Nij
- Nik)2]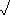 [(15-12)2
+ (9-8)2] =
[(15-12)2
+ (9-8)2] = 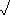 10 = 3.16
10 = 3.16